Study on the Anti-Colorectal Cancer Mechanism of Jianpi Jiedu Decoction Based on Network Pharmacology
-
摘要:
目的 基于网络药理学和分子对接研究健脾解毒方的抗结直肠癌作用机制。 方法 运用TCMSP、TCMID、ETCM数据库预测健脾解毒方有效成分及其作用靶点, 利用GeneCards数据库检索肿瘤相关靶点, 并进行药物-疾病靶点匹配。将共同靶点导入Cytoscape 3.7.2构建PPI网络并进行网络拓扑分析筛选关键靶点。通过R 3.6.3软件对共同靶点进行GO富集分析及KEGG通路富集分析。利用AutoDock平台进行分子对接, 预测有效成分与关键靶点的结合度, 并对关键靶点进行实验验证。 结果 共筛选出健脾解毒方有效成分24种, 肿瘤相关靶点86个, 其中关键靶点为PTGS2、HSP90AA1、PRSS1, 且与主要有效成分槲皮素、山柰酚均具有良好的结合活性。GO富集分析和KEGG通路富集分析提示健脾解毒方可能通过PI3K-Akt信号通路、MAPK信号通路发挥抗结直肠癌作用。实验验证发现健脾解毒方能够下调PTGS2、p38MAPK蛋白及mRNA表达, 而沉默PTGS2基因后健脾解毒方对下游基因p38MAPK的表达无明显调控作用, 且对结肠癌细胞转移能力无明显影响。 结论 健脾解毒方能够抑制结肠癌转移, 其机制与PTGS2介导的p38MAPK信号通路相关。 Abstract:OBJECTIVE To explore the anti-tumor mechanism of Jianpi Jiedu Decoction (JPJDD) based on network pharmacology and molecular docking. METHODS TCMSP, TCMID and ETCM databases were used to predict the active ingredients and the targets of JPJDD, and GeneCards database was used to search tumor related targets and match the targets above. The common targets were imported into Cytoscape 3.7.2 to construct the PPI network and select the key targets by network topology analysis. GO enrichment analysis and KEGG pathway enrichment analysis were performed by R 3.6.3 software. Finally the Autodock 2.4 was used for molecular docking to predict the binding degree of the active ingredients to target genes. In addition, the key targets were verified by cell experiments. RESULTS 24 kinds of active ingredients and 86 tumor related targets were identified, including the key targets of PTGS2, HSP90AA1 and PRSS1, which were obtained by PPI network topology analysis. The molecular docking showed that all of the key targets had good binding activity with quercetin and kaempferol. GO enrichment analysis and KEGG pathway enrichment analysis suggested that JPJDD may play the role of anti-tumor through PI3K/Akt signal pathway and MAPK signal pathway. The results showed that JPJDD inhibited both the protein and mRNA expressions of PTGS2 and p38MAPK, but after silencing PTGS2 gene, the expression of downstream gene p38MAPK and the effect on colorectal cancer cells metastasis had no obvious response to JPJDD. CONCLUSION JPJDD can inhibit the metastasis of colorectal cancer via PTGS2 mediated p38MAPK signaling pathway. -
Key words:
- Jianpi Jiedu Decoction /
- network pharmacology /
- colorectal cancer /
- PTGS2 /
- p38MAPK
-
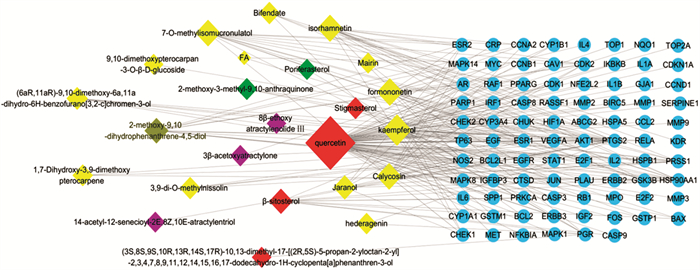
表 1 健脾解毒方的有效化学成分
Table 1. Active ingredients of Jianpi Jiedu Decoction (JPJDD)
来源 有效成分 OB DL 节点度 黄芪 Mairin 55.38 0.78 1 Jaranol 50.83 0.29 1 Hederagenin 36.91 0.75 4 Isorhamnetin 49.60 0.31 14 3, 9-di-O-Methylnissolin 53.74 0.48 4 7-O-Methylisomucronulatol 74.69 0.30 13 9, 10-Dimethoxypterocarpan-3-O-β-D-glucoside 36.74 0.92 1 (6aR, 11aR)-9, 10-Dimethoxy-6a, 11a-dihydro-6H-benzofurano[3, 2-c]chromen-3-ol 64.26 0.42 5 Bifendate 31.10 0.67 1 Formononetin 69.67 0.21 15 Calycosin 47.75 0.24 13 Kaempferol 41.88 0.24 23 FA 68.96 0.71 2 Isomucronulatol-7, 2'-di-O-glucosiole 49.28 0.62 0 1, 7-Dihydroxy-3, 9-dimethoxy pterocarpene 39.05 0.48 3 白术 14-Acetyl-12-senecioyl-2E, 8Z, 10E-atractylentriol 60.31 0.31 1 3β-Acetoxyatractylone 54.07 0.22 2 8β-Ethoxy atractylenolide Ⅲ 35.95 0.21 1 白花蛇舌草 Poriferasterol 43.83 0.76 1 2-Methoxy-3-methyl-9, 10-anthraquinone 37.83 0.21 3 山慈菇 2-Methoxy-9, 10-dihydrophenanthrene-4, 5-diol 44.97 0.18 11 共有成分 Stigmasterol 43.83 0.76 3 β-Sitosterol 36.91 0.75 10 Quercetin 46.43 0.28 72 (3S, 8S, 9S, 10R, 13R, 14S, 17R)-10, 13-Dimethyl-17-[(2R, 5S)-5-propan-2-yloctan-2-yl]-2, 3, 4, 7, 8, 9, 11, 12, 14, 15, 16, 17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol 36.23 0.78 1 表 2 健脾解毒方活性成分与关键靶点的分子对接结果
Table 2. The dock results of the active ingredients of JPJDD and the key targets
成分 结合自由能/(kJ·mol-1) PTGS2 PRSS1 HSP90AA1 槲皮素 -26.417 6 -27.588 0 -25.748 8 山柰酚 -26.166 8 -26.877 4 -27.755 2 -
[1] HOPKINS AL. Network pharmacology: The next paradigm in drug discovery[J]. Nat Chem Biol, 2008, 4(11): 682-690. doi: 10.1038/nchembio.118 [2] KIBBLE M, SAARINEN N, TANG J, et al. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products[J]. Nat Prod Rep, 2015, 32(8): 1249-1266. doi: 10.1039/C5NP00005J [3] YUAN HD, MA QQ, CUI HY, et al. How can synergism of traditional medicines benefit from network pharmacology?[J]. Molecules, 2017, 22(7): 1135. doi: 10.3390/molecules22071135 [4] MUHAMMAD J, KHAN A, ALI A, et al. Network pharmacology: Exploring the resources and methodologies[J]. Curr Top Med Chem, 2018, 18(12): 949-964. doi: 10.2174/1568026618666180330141351 [5] RU J, LI P, WANG J, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines[J]. J Cheminform, 2014, 6: 13. doi: 10.1186/1758-2946-6-13 [6] XUE R, FANG Z, ZHANG M, et al. TCMID: Traditional Chinese Medicine integrative database for herb molecular mechanism analysis[J]. Nucleic Acids Res, 2013, 41: D1089-D1095. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3531123/pdf/gks1100.pdf [7] XU HY, ZHANG YQ, LIU ZM, et al. ETCM: an encyclopaedia of traditional Chinese medicine[J]. Nucleic Acids Res, 2019, 47(d1): D976-D982. doi: 10.1093/nar/gky987 [8] WISHART DS, FEUNANG YD, GUO AC, et al. DrugBank 5.0: A major update to the DrugBank database for 2018[J]. Nucleic Acids Res, 2018, 46(d1): D1074-D1082. doi: 10.1093/nar/gkx1037 [9] REBHAN M, CHALIFA-CASPI V, PRILUSKY J, et al. GeneCards: A novel functional genomics compendium with automated data mining and query reformulation support[J]. Bioinformatics, 1998, 14(8): 656-664. doi: 10.1093/bioinformatics/14.8.656 [10] TIPPMANN S. Programming tools: Adventures with R[J]. Nature, 2015, 517(7532): 109-110. doi: 10.1038/517109a [11] SHANNON P, MARKIEL A, OZIER O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks[J]. Genome Res, 2003, 13(11): 2498-2504. doi: 10.1101/gr.1239303 [12] MARTIN A, OCHAGAVIA ME, RABASA LC, et al. BisoGenet: A new tool for gene network building, visualization and analysis[J]. BMC Bioinformatics, 2010, 11: 91. doi: 10.1186/1471-2105-11-91 [13] ZEILEIS A, HORNIK K, MURRELL P. Escaping RGBland: Selecting colors for statistical graphics[J]. Comput Stat Data Anal, 2009, 53(9): 3259-3270. doi: 10.1016/j.csda.2008.11.033 [14] LAWRENCE M, HUBER W, PAGES H, et al. Software for computing and annotating genomic ranges[J]. PLoS Comput Biol, 2013, 9(8): e1003118. doi: 10.1371/journal.pcbi.1003118 [15] WICKHAM H. ggplot2: Elegant Graphics for Data Analysis[M]. NewYork: Springer-Verlag, 2016: 8. [16] HUBER W, CAREY VJ, GENTLEMAN R, et al. Orchestrating high-throughput genomic analysis with Bioconductor[J]. Nature Methods, 2015, 12(2): 115-121. doi: 10.1038/nmeth.3252 [17] BERMAN HM, WESTBROOK J, FENG Z, et al. The protein data bank[J]. Nucleic Acids Res, 2000, 28(1): 235-242. doi: 10.1093/nar/28.1.235 [18] MORRIS GM, HUEY R, LINDSTROM W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility[J]. J Comput Chem, 2009, 30(16): 2785-2791. doi: 10.1002/jcc.21256 [19] JASON G, PAIARDINI A. PyMod 3: A complete suite for structural bioinformatics in PyMOL[J]. Bioinformatics, 2021, 37(10): 1471-1472. doi: 10.1093/bioinformatics/btaa849 [20] ALMATROODI SA, ALSAHLI MA, ALMATROUDI A, et al. Potential therapeutic targets of quercetin, a plant flavonol, and its role in the therapy of various types of cancer through the modulation of various cell signaling pathways[J]. Molecules, 2021, 26(5): 1315. doi: 10.3390/molecules26051315 [21] LI QY, WEI LH, LIN S, et al. Synergistic effect of kaempferol and 5-fluorouracil on the growth of colorectal cancer cells by regulating the PI3K/Akt signaling pathway[J]. Mol Med Rep, 2019, 20(1): 728-734. [22] WANG X, YANG Y, AN Y, et al. The mechanism of anticancer action and potential clinical use of kaempferol in the treatment of breast cancer[J]. Biomed Pharmacother, 2019, 117: 109086. doi: 10.1016/j.biopha.2019.109086 [23] BROWN JR, DUBOIS RN. COX-2: A molecular target for colorectal cancer prevention[J]. J Clin Oncol, 2005, 23(12): 2840-2855. doi: 10.1200/JCO.2005.09.051 [24] CHAN AT, OGINO S, FUCHS CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2[J]. N Engl J Med, 2007, 356(21): 2131-2142. doi: 10.1056/NEJMoa067208 [25] CUENDA A, ROUSSEAU S. p38 MAP-kinases pathway regulation, function and role in human diseases[J]. Biochim Biophys Acta, 2007, 1773(8): 1358-1375. doi: 10.1016/j.bbamcr.2007.03.010 -




 下载:
下载: